Our research focuses on studying human brain transcriptome using next-generation sequencing, in particular RNA-Seq technique. We are interested in region-specific gene expression profiles in a healthy human brain as well as perturbation of the transcriptome as a result of neurodegeneration in multiple system atrophy (MSA) and Alzheimer's disease (AD).
Multiple System Atrophy
MSA is a distinct member of the group of neurodegenerative diseases called α-synucleinopathies whereby the fibrillar protein α-synuclein aggregates in oligodendroglia. Clinical symptoms of MSA comprise autonomic failure in combination with parkinsonism or cerebellar ataxia. Although well defined clinically the molecular pathophysiology of MSA has been barely investigated. In particular, there have been no systematic studies of the perturbation of the brain transcriptome during the onset and persistence of this disease. Interestingly measurements of the α-synuclein gene (SNCA) expression in MSA brain tissue did not revealed overexpression of this gene in the oligodendrocytes. It therefore became clear that other genes and gene networks, both directly, as non-coding RNAs, or through their protein products contribute to accelerated translation and accumulation of the alpha-synuclein protein in the white matter of the brain.
Alzheimer's Disease
AD is the most common cause of dementia that mainly affects individuals over the age of 60, with a risk that increases steadily in older ages. It is characterised by a complex progression of neurodegeneration resulting in impairments in memory and other cognitive processes, as well as the presence of noncognitive symptoms. Histopathologically AD is featured by the presence of β-amyloid-containing senile plaques due to irregular amyloid precursor protein (APP) processing and hyperphosphorylated tau-containing neurofibrillary tangles. Transcriptome analysis in AD has been mostly performed using transgenic animals and patient-derived cell lines. Moreover gene expression studies, using microarrays, in post-mortem brain tissue generated mostly discordant results. Emerging concepts of pervasive transcription and the role of non-coding RNA in regulation of cellular physiology brought a new perspective for investigation of AD-specific pathomechanism of neurodegeneration.
Our Goals
The main aim of our research is to identify transcriptional markers specific for particular structures and regions of the normal human brain using genome-wide analysis approaches such as transcriptome sequencing. Moreover, using RNA-Seq and bioinformatics tools we aim to identify transcriptional isoforms of which expression is influenced by neurodegenerative process in MSA and AD. The scope of our analysis covers both protein-coding and non-coding RNAs, with particular focus on long intergenic non-coding RNAs (lincRNAs).

Figure 1: Brain explorer image from Allen Brain Atlas showing the transcription levels of SNCA througout the brain. Red represents up-regulation of SNCA, green represents down-regulation SNCA.
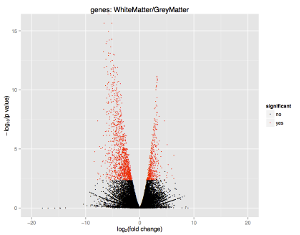
Figure 2: Volcano plot of gene expression in white matter (WM) and grey matter (GM) from the human brain. Each point represents one gene, those genes with a negative fold-change were up-regulated in GM and those genes with a positive fold-change were up-regulated in WM. Red represents those genes that are differenetially expressed.